当前位置:
X-MOL 学术
›
Mol. Biosyst.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Global DNA dynamics of 8-oxoguanine repair by human OGG1 revealed by stopped-flow kinetics and molecular dynamics simulation
Molecular BioSystems Pub Date : 2017-07-14 00:00:00 , DOI: 10.1039/c7mb00343a M. V. Lukina 1, 2, 3, 4, 5 , V. V. Koval 1, 2, 3, 4, 5 , A. A. Lomzov 1, 2, 3, 4, 5 , D. O. Zharkov 1, 2, 3, 4, 5 , O. S. Fedorova 1, 2, 3, 4, 5
Molecular BioSystems Pub Date : 2017-07-14 00:00:00 , DOI: 10.1039/c7mb00343a M. V. Lukina 1, 2, 3, 4, 5 , V. V. Koval 1, 2, 3, 4, 5 , A. A. Lomzov 1, 2, 3, 4, 5 , D. O. Zharkov 1, 2, 3, 4, 5 , O. S. Fedorova 1, 2, 3, 4, 5
Affiliation
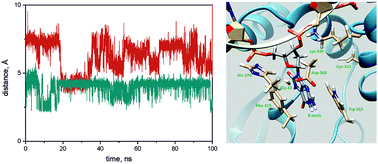
|
The toxic action of different endogenous and exogenous agents leads to damage in genomic DNA. 8-Oxoguanine is one of the most often generated and highly mutagenic oxidative forms of damage in DNA. Normally, in human cells it is promptly removed by 8-oxoguanine-DNA-glycosylase hOGG1, the key DNA-repair enzyme. An association between the accumulation of oxidized guanine and an increased risk of harmful processes in organisms was already found. However, the detailed mechanism of damaged base recognition and removal is still unclear. To clarify the role of active site amino acids in the damaged base coordination and to reveal the elementary steps in the overall enzymatic process we investigated hOGG1 mutant forms with substituted amino acid residues in the enzyme base-binding pocket. Replacing the functional groups of the enzyme active site allowed us to change the rates of the individual steps of the enzymatic reaction. To gain further insight into the mechanism of hOGG1 catalysis a detailed pre-steady state kinetic study of this enzymatic process was carried out using the stopped-flow approach. The changes in the DNA structure after mixing with enzymes were followed by recording the FRET signal using Cy3/Cy5 labels in DNA substrates in the time range from milliseconds to hundreds of seconds. DNA duplexes containing non-damaged DNA, 8-oxoG, or an AP-site or its unreactive synthetic analogue were used as DNA-substrates. The kinetic parameters of DNA binding and damage processing were obtained for the mutant forms and for WT hOGG1. The analyses of fluorescence traces provided information about the DNA dynamics during damage recognition and removal. The kinetic study for the mutant forms revealed that all introduced substitutions reduced the efficiency of the hOGG1 activity; however, they played pivotal roles at certain elementary stages identified during the study. Taken together, our results gave the opportunity to restore the role of substituted amino acids and main “damaged base–amino acid” contacts, which provide an important link in the understanding the mechanism of the DNA repair process catalyzed by hOGG1.
中文翻译:

停止流动力学和分子动力学模拟揭示了人类OGG1修复8-氧鸟嘌呤的全球DNA动力学
不同的内源性和外源性物质的毒性作用会导致基因组DNA的破坏。8-氧鸟嘌呤是DNA损伤中最常见的一种,并且是高度诱变的氧化形式。通常,在人类细胞中,它会被8-氧鸟嘌呤-DNA-糖基化酶hOGG1(关键的DNA修复酶)迅速清除。已经发现氧化鸟嘌呤的积累与生物体内有害过程的风险增加之间存在关联。但是,受损碱基识别和清除的详细机制仍不清楚。为了阐明活性位点氨基酸在受损的碱基配位中的作用并揭示整个酶促过程中的基本步骤,我们研究了在酶碱基结合袋中带有取代氨基酸残基的hOGG1突变体形式。取代酶活性位点的官能团使我们能够改变酶促反应各个步骤的速率。为了进一步了解hOGG1催化的机理,使用了停流方法对该酶过程进行了详细的稳态前动力学研究。与酶混合后,DNA结构发生变化,然后使用Cy3 / Cy5标签在DNA底物中记录FRET信号,时间范围为几毫秒到几百秒。包含未损坏的DNA,8-oxoG或AP位点或其无反应的合成类似物的DNA双链体被用作DNA底物。获得了突变形式和WT hOGG1的DNA结合和损伤过程的动力学参数。荧光迹线的分析提供了有关损伤识别和清除过程中DNA动态的信息。突变体形式的动力学研究表明,所有引入的取代均降低了hOGG1活性的效率。但是,他们在研究过程中确定的某些基本阶段发挥了关键作用。综上所述,我们的结果提供了恢复取代氨基酸和主要“受损碱基-氨基酸”接触的作用的机会,这在理解hOGG1催化的DNA修复过程的机理中提供了重要的联系。
更新日期:2017-08-03
中文翻译:

停止流动力学和分子动力学模拟揭示了人类OGG1修复8-氧鸟嘌呤的全球DNA动力学
不同的内源性和外源性物质的毒性作用会导致基因组DNA的破坏。8-氧鸟嘌呤是DNA损伤中最常见的一种,并且是高度诱变的氧化形式。通常,在人类细胞中,它会被8-氧鸟嘌呤-DNA-糖基化酶hOGG1(关键的DNA修复酶)迅速清除。已经发现氧化鸟嘌呤的积累与生物体内有害过程的风险增加之间存在关联。但是,受损碱基识别和清除的详细机制仍不清楚。为了阐明活性位点氨基酸在受损的碱基配位中的作用并揭示整个酶促过程中的基本步骤,我们研究了在酶碱基结合袋中带有取代氨基酸残基的hOGG1突变体形式。取代酶活性位点的官能团使我们能够改变酶促反应各个步骤的速率。为了进一步了解hOGG1催化的机理,使用了停流方法对该酶过程进行了详细的稳态前动力学研究。与酶混合后,DNA结构发生变化,然后使用Cy3 / Cy5标签在DNA底物中记录FRET信号,时间范围为几毫秒到几百秒。包含未损坏的DNA,8-oxoG或AP位点或其无反应的合成类似物的DNA双链体被用作DNA底物。获得了突变形式和WT hOGG1的DNA结合和损伤过程的动力学参数。荧光迹线的分析提供了有关损伤识别和清除过程中DNA动态的信息。突变体形式的动力学研究表明,所有引入的取代均降低了hOGG1活性的效率。但是,他们在研究过程中确定的某些基本阶段发挥了关键作用。综上所述,我们的结果提供了恢复取代氨基酸和主要“受损碱基-氨基酸”接触的作用的机会,这在理解hOGG1催化的DNA修复过程的机理中提供了重要的联系。


























 京公网安备 11010802027423号
京公网安备 11010802027423号