当前位置:
X-MOL 学术
›
J. Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Large-Scale Analysis of Hydrogen Bond Interaction Patterns in Protein–Ligand Interfaces
Journal of Medicinal Chemistry ( IF 7.3 ) Pub Date : 2017-05-12 00:00:00 , DOI: 10.1021/acs.jmedchem.7b00101 Eva Nittinger 1 , Therese Inhester 1 , Stefan Bietz 1 , Agnes Meyder 1 , Karen T. Schomburg 1 , Gudrun Lange 2 , Robert Klein 2 , Matthias Rarey 1
Journal of Medicinal Chemistry ( IF 7.3 ) Pub Date : 2017-05-12 00:00:00 , DOI: 10.1021/acs.jmedchem.7b00101 Eva Nittinger 1 , Therese Inhester 1 , Stefan Bietz 1 , Agnes Meyder 1 , Karen T. Schomburg 1 , Gudrun Lange 2 , Robert Klein 2 , Matthias Rarey 1
Affiliation
|
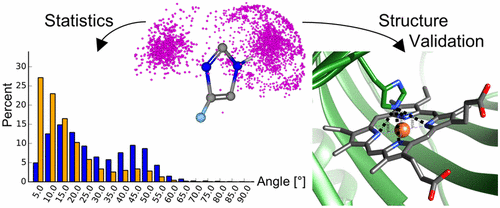
Protein–ligand interactions are the fundamental basis for molecular design in pharmaceutical research, biocatalysis, and agrochemical development. Especially hydrogen bonds are known to have special geometric requirements and therefore deserve a detailed analysis. In modeling approaches a more general description of hydrogen bond geometries, using distance and directionality, is applied. A first study of their geometries was performed based on 15 protein structures in 1982. Currently there are about 95 000 protein–ligand structures available in the PDB, providing a solid foundation for a new large-scale statistical analysis. Here, we report a comprehensive investigation of geometric and functional properties of hydrogen bonds. Out of 22 defined functional groups, eight are fully in accordance with theoretical predictions while 14 show variations from expected values. On the basis of these results, we derived interaction geometries to improve current computational models. It is expected that these observations will be useful in designing new chemical structures for biological applications.
中文翻译:

蛋白质-配体界面中氢键相互作用模式的大规模分析
蛋白质-配体相互作用是药物研究,生物催化和农药开发中分子设计的基本基础。众所周知,尤其是氢键具有特殊的几何要求,因此值得详细分析。在建模方法中,使用距离和方向性对氢键几何形状进行了更一般的描述。1982年,基于15种蛋白质结构对它们的几何形状进行了首次研究。目前,PDB中大约有95 000种蛋白质-配体结构可用,这为新的大规模统计分析奠定了坚实的基础。在这里,我们报告了氢键的几何和功能特性的全面研究。在22个定义的功能组中,8个完全符合理论预测,而14个显示与期望值的差异。基于这些结果,我们推导了交互几何结构以改进当前的计算模型。预期这些观察将对设计用于生物学应用的新化学结构有用。
更新日期:2017-05-12
中文翻译:

蛋白质-配体界面中氢键相互作用模式的大规模分析
蛋白质-配体相互作用是药物研究,生物催化和农药开发中分子设计的基本基础。众所周知,尤其是氢键具有特殊的几何要求,因此值得详细分析。在建模方法中,使用距离和方向性对氢键几何形状进行了更一般的描述。1982年,基于15种蛋白质结构对它们的几何形状进行了首次研究。目前,PDB中大约有95 000种蛋白质-配体结构可用,这为新的大规模统计分析奠定了坚实的基础。在这里,我们报告了氢键的几何和功能特性的全面研究。在22个定义的功能组中,8个完全符合理论预测,而14个显示与期望值的差异。基于这些结果,我们推导了交互几何结构以改进当前的计算模型。预期这些观察将对设计用于生物学应用的新化学结构有用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号